From NutriBench to Taralli: How far can you take a prompt?
There's something very funny about the current Machine Learning and AI landscape. If you're in the field you probably heard about it. "Vibes" they call it. When someone wants to test something out, they conduct a "vibe test".
I call bullshit. How are you supposed to improve something if you can't even measure it? How do you know you didn't just make it worse?
In the past month, I've been working on Taralli. And I'd like to show you how to incrementally improve an LLM based system.
But first, let's take a little side quest.
NutriBench: Evaluating LLMs on nutrition estimation
While browsing arXiv the other day I stumbled upon NutriBench. This is exciting - I thought to myself - it looks like someone else is also looking into this problem. NutriBench is a research project launched by the University of California that looks to quantify exactly Taralli's problem: "How good are Large Language Models at estimating the nutritional content of foods?".
The team created a dataset for ~12K meal descriptions from two data sources (WWEIA and FAO/WHO). Every row in the dataset is a meal description with the corresponding nutritional details (carbs, protein, fat, and energy). To answer the question, they tested 4 different prompting techniques and 12 different LLMs. The prompting techniques were the following:
- Base: A simple prompt with some few-shot examples.
- Chain-of-Thought (CoT): A slightly more complex prompt with some reasoning steps to "force" the model to reason.
- Retrieval-Augmented Generation (RAG): A system that uses a retrieval database (which they called Retri-DB), to aid the LLM in estimating carbs.
- RAG+CoT: A mix of the former two.
If you are curious about the prompts, click here.
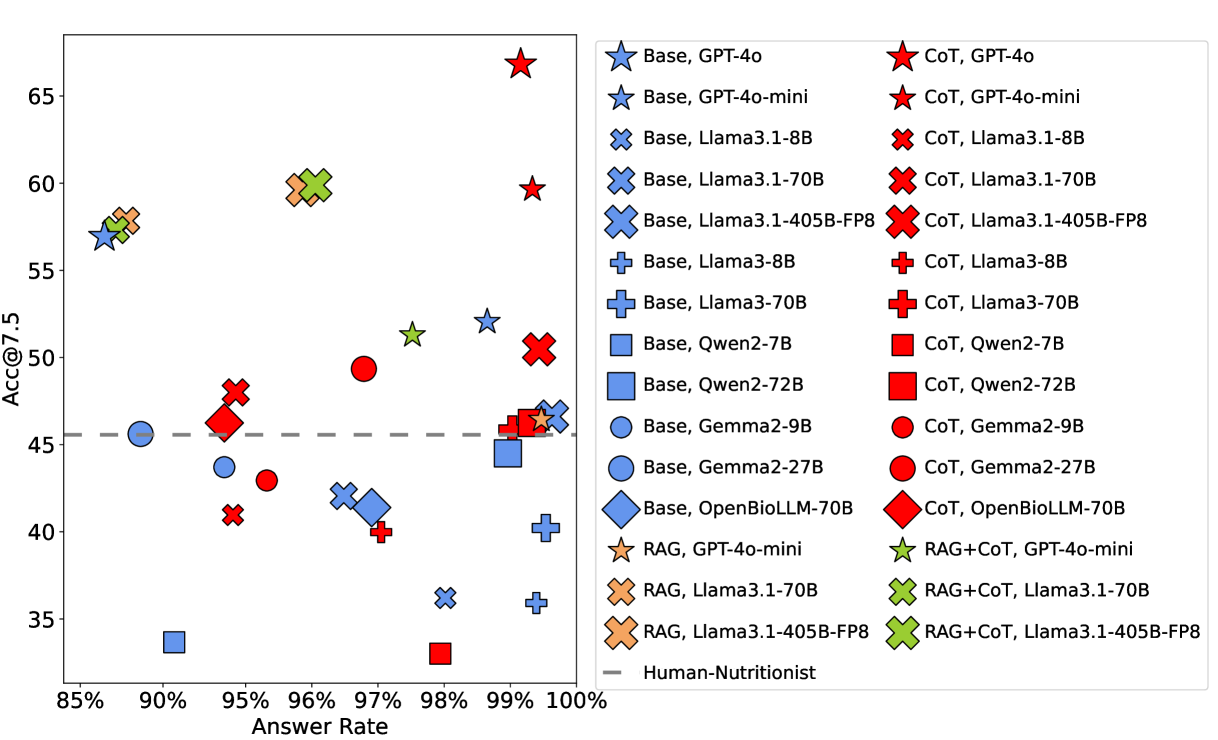
You're probably wondering: okay, but how good are the models at this? The answer is a resounding MEH. The best performer is GPT-4o with a Chain-of-Thought prompt. It responds to ~99% of prompts (models can refuse to answer) and achieves an [email protected] of 66.82%1. In other words, its carb predictions fall within ±7.5g of the actual value about two-thirds of the time.
What if we could use the NutriBench dataset to improve Taralli?
Improving Taralli's nutritional estimation
Here's the notebook with the entire process
The first thing, as always, is to create good dataset. I created a new dataset of 107 examples of food descriptions and their corresponding total calories2. I used the second version of the NutriBench dataset on Hugging Face. I mixed in that dataset with some other examples from a previous golden dataset.
Expand to see some examples of the dataset
[
{
"input": "For dinner, I'm having 25 grams of bread, 150 grams of chicken wings, and a 250-gram mixed vegetable salad.",
"total_calories": 633.0,
"food_groups": [
"grain",
"meat and alternatives",
"vegetable"
],
"source": "nutribench"
},
{
"input": "I enjoyed 200 grams of tea with sugar along with 230 grams of coconut milk rice for breakfast.",
"total_calories": 558.0,
"food_groups": [
"grain",
"fruit"
],
"source": "nutribench"
},
{
"input": "stracciatella with confit grapes and 2 little focaccia pieces",
"total_calories": 350.0,
"food_groups": [
"fruit",
"dairy",
"grain"
],
"source": "golden_dataset"
}
]
Now I needed to design an evaluation metric. For NutriBench they used accuracy of carb prediction at ±7.5 grams. In our case, I'm more interested in the accuracy calorie prediction. One of the nice things about DSPy is that it forces you to write down your evaluation metric explicitly. Below is the evaluation metric I used: In short, we want the predicted calories to be within 10% of the ground truth calories. Which means our metric will then be Accuracy@10%.
def eval_metric(
example, pred, trace=None, pred_name=None, pred_trace=None
) -> dspy.Prediction:
# see notebook for more detailed metric (this is simplified)
total_calories_example = example.total_calories
total_calories_predicted = na.total_calories()
within_threshold = abs(total_calories_example - total_calories_predicted) <= abs(
0.1 * total_calories_example
)
if not within_threshold:
score = 0
feedback_text = f"INCORRECT: Your answer was not within 10% of the correct answer (yours: {total_calories_predicted}, correct: {total_calories_example})"
return dspy.Prediction(score=score, feedback=feedback_text)
feedback_text = f"CORRECT: Your answer was within 10% of the correct answer (yours: {total_calories_predicted}, correct: {total_calories_example})"
score = 1
return dspy.Prediction(score=score, feedback=feedback_text)
Here are the different prompts I tested:
- Vanilla: Zero shot prompt using the DSPy format.
- Bootstrap few shot golden only: Using the golden dataset I previously collected. This is what was running in production for Taralli. BootstrapFewShotWithRandomSearch
- Bootstrap few mixed: Uses a dataset that is a mix of my data and the NutriBench data. BootstrapFewShotWithRandomSearch
- MIPROv2: Multiprompt Instruction PRoposal Optimizer Version 2, a prompt optimizer capable of optimizing both the instructions (e.g., the prompt) and the few shot examples. MIPROv2
- GEPA: The new kid on the block for prompt optimizers. The nice thing about GEPA is that you can also give it some textual feedback on the incorrect predictions, and it will use it. GEPA
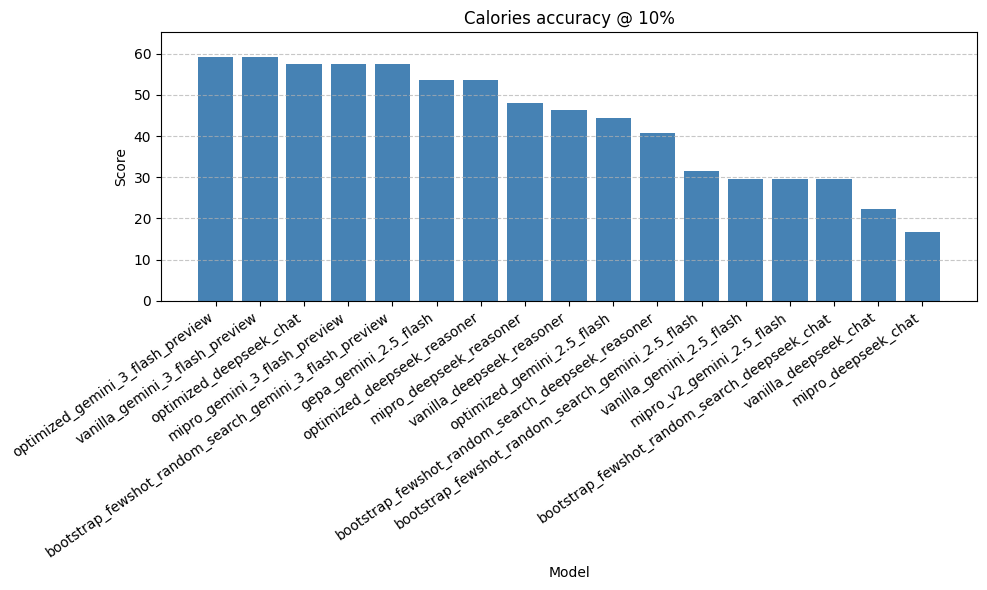
As for models, I chose some that were cheap and fast - some closed (Gemini) and other Open (DeepSeek). I decided to test, Gemini 2.5 Flash (what I am currently using in production), DeepSeech v3.2 with thinking on and off, and Google's new Gemini 3 Flash.
Some thoughts on the results:
- The best performing model is Gemini 3 Flash with a set of 16 examples in the prompt. It achieves a score of around 60%. Similar to NutriBench, although the problems are slightly different. Here's an example prediction.
- The GEPA optimization came up with a prompt itself. When using that prompt with Gemini 2.5 Flash it performs respectably well. However, the GEPA prompt actually fails to provide the correct response format when used with any other model. In other words, the prompt was overfit to the model it was trained on. As a result, you only see one GEPA score in the results graph.
- The few-shot approach is the most reliable one. It is model agnostic, performs well, and follows the styles of examples in a faithful manner.
So, I decided to update Taralli to use Gemini 3 Flash with the few-shot approach. This approach is ~15% more accurate when compared to the old version, which was running on Gemini 2.5 Flash, with the exact same optimizer. In conclusion, all I changed was a model string.
Here's a snippet taken from the API. Since I'm using OpenRouter, I can also define our 2nd best performing model as a back up, just like so:
LM = dspy.LM(
"openrouter/google/gemini-3-flash-preview:nitro",
temperature=0.0,
extra_body={
"reasoning": {"enabled": False},
"models": ["deepseek/deepseek-v3.2:nitro"],
},
)
Running on the edge
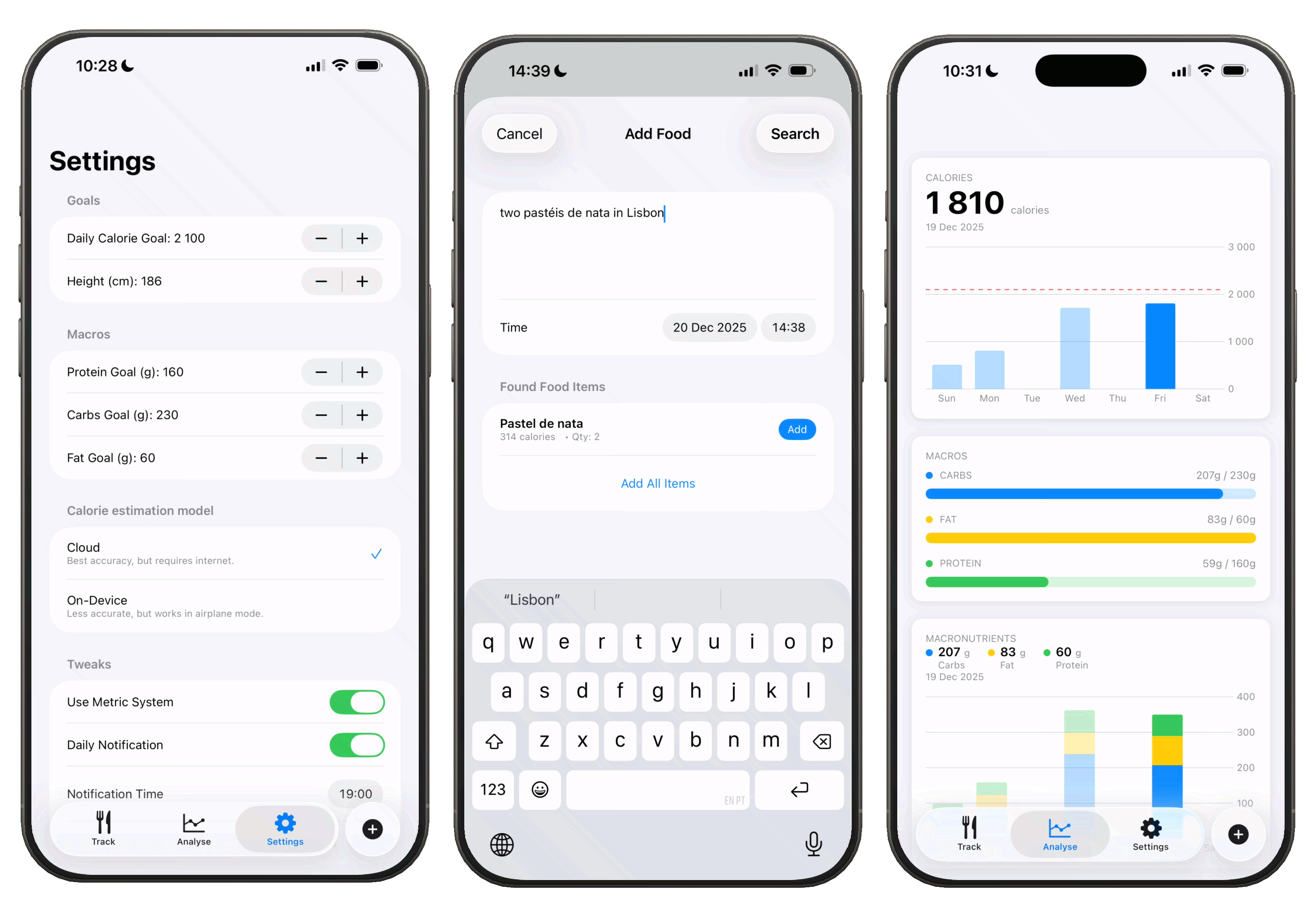
I've updated Taralli to use Apple's new Liquid Glass design for iOS 26. That was pretty simple. It's a 5 file SwitfUI app with around 4.5 MB. You can now also set periodic reminders so that you don't forget to track.
One of the best new features is the ability to use Taralli completely offline with an on-device LLM for nutritional analysis. The code snippet below transforms a DSPy optimized program into OpenAI-compatible messages. I created an endpoint that takes a food_description and returns these messages for on-device inference. The iOS app calls this endpoint, receives the populated template, and uses it with the on-device model. As a backup, we can bundle the template directly in the app to skip the API call entirely. Long story short, Taralli now works on airplane (mode).
@lru_cache(maxsize=1)
def get_classifier_template() -> t.List[dict[str, str]]:
program = get_classifier()
adapter = dspy.ChatAdapter()
openai_messages_format = {
name: adapter.format(
p.signature, # type: ignore
demos=p.demos,
inputs={k: f"{{{k}}}" for k in p.signature.input_fields}, # type: ignore
)
for name, p in program.named_predictors()
}["self"]
return openai_messages_format
# returns [{"role": "system", "content": feajfeafl}, {"role..}]
Final summary
- Hill climbing NutriBench: The best performing model/prompt combo in my experiments hit a 60% Accuracy @ 10%. The task is slightly different than NutriBench - we also predict fat, protein, and carbs for example. However, I'm still interested in exploring how we could possibly hill climb that dataset. Could we get to 90%? Should we try a larger model? More thinking? Web browsing? How much of a difference would it make?
- Training a model: We could obviously also train a model on this task. The prompt optimizations will only get you so far, but it would also be very interesting to see how much a fine-tuned model would perform. To be completely honest, I still believe that in order to get the best performance possible, we would probably need some sort of connection to the outside world (web search, RAG, or the likes).
- Few shot wins: From several experiments with Taralli, and a couple of other projects, something has become more and more obvious: few-shot often wins. Not only when it relates to accuracy, but also specifying the correct format for the output that you are looking for. If you are dealing with a complicated task - try squeezing in some good examples of model behaviour.
- Vibes are useless: You can't improve something you can't measure. Whenever someone tells me "the vibes are off," I call bullshit. Off? By how much? Can we put a number to it? Whenever you are improving a system, try to be scientific about it. Measure first, understand how the current system is performing, try to improve it, and then measure again.